Publications
39. Howland K, Brock A. (2023) Cellular barcoding tracks heterogeneous clones through selective pressures and phenotypic transitions. Trends Cancer. https://doi.org/10.1016/j.trecan.2023.03.008
38. Adler F, Anderson A.R.A., Bhushan A, Bogdan P, Bravo-Cordero JJ, Brock A, Chen Y, Cukierman E, DelGiorno KE, Denis GV, Ferrall-Fairbanks M, Gartner Z, Germain R, Gordon DM, Hunter G, Jolly MK, Karacosta LG, Mythreye K, Katira P, Kulkarni RP, Kutys ML, Lander AD, Laughney AM, Levine H, Lou E; Lowenstein PR, Masters KS, Pe'er D, Peyton SR, Platt MO, Purvis JE, Quon G, Richer JK, Riddle NC, Rodriguez A, Snyder JC, Szeto GL, Tomlin CJ, Yanai I, Zervantonakis I, Dueck HR. (2023) Modeling collective cell behavior in cancer: perspectives from an interdisciplinary conversation. Cell Systems. https://doi.org/10.1016/j.cels.2023.03.002
37. Howard G, Jost T, Yankeelov TE, Brock A. (2022) Quantification of long-term doxorubicin response dynamics in breast cancer cell lines to direct treatment schedules. PLOS Comp Biol 18 (3), e1009104. doi: https://doi.org/10.1101/2021.05.24.445407
36. Cotner M, Meng S, De Santiago C, Gardener A, Jost T, Brock A. (2022) Integration of quantitative methods and mathematical approaches for the modeling of cell proliferation dynamics. Amer. J. Cell Phys, 234(2), C247-262. doi.org/10.1152/ajpcell.00185.2022
35. Gardner A*, Morgan D*, Al’Khafaji A, Brock A. (2022) Functionalized Lineage Tracing for the Study and Manipulation of Heterogeneous Cell Populations. Biomedical Engineering Technologies. Methods in Molecular Biology, vol 2394. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-1811-0_8
34. Morgan D, Jost TA, DeSantiago C, Brock, A. (2021) Applications of high-resolution clone tracking technologies in cancer. Curr Opinion in Biomed Eng,100317.
33. Yang J, Virostko J, Hormuth DA, Liu J, Brock A, Kowalski J, Yankeelov TE (2021). An experimental-mathematical approach to predict tumor cell growth as a function of glucose availability in breast cancer cell lines. PloS one, 16 (7), e0240765.
32. Gutierrez C*, Al’Khafaji AM*, Brenner E*, Johnson KE, Gohil SH, Lin Z, Knisbacher BA, Durrett RE, Li S, Parvin S, Biran A, Zhang W, Rassenti L, Kipps TJ, Livak KJ, Neuberg D, Letai A, Getz G, Wu CJ, Brock A. (2021) Multifunctional barcoding with ClonMapper enables high-resolution study of clonal dynamics during tumor evolution and treatment. Nat Cancer, 2, 758–772. https://doi.org/10.1038/s43018-021-00222-8.
31. Liu J, Hormuth DA II, Davis T, Yang J, McKenna MT, Jarrett AM, Enderling H, Brock A, Yankeelov TE (2021) A time-resolved experimental–mathematical model for predicting the response of glioma cells to single-dose radiation therapy, Integrat Biol,13:7 167–183. https://doi.org/10.1093/intbio/zyab010
30. Kazerouni AS, Gadde M, Gardner A, Hormuth DA, Jarrett AM, Johnson KE, Lima EABF, Lorenzo G, Phillips C, Brock A, Yankeelov TE (2020). Integrating quantitative assays with biologically-based mathematical modeling for predictive oncology. iScience 101807. https://doi.org/10.1016/j.isci.2020.101807 3
29. Johnson K, Howard G, Morgan D, Brenner E, Gardner A, Durrett R, Mo W, Al’Khafaji A, Sontag E, Jarrett A, Yankeelov T, Brock A (2020) Integrating multimodal data sets into a mathematical framework to describe and predict therapeutic resistance in cancer. Phys Biol. 18 016001. https://doi.org/10.1088/1478-3975/abb09c.
28. Phillips CM, Lima EABF, Woodall RT, Brock A, Yankeelov TE (2020) A hybrid model of tumor growth and angiogenesis: In silico experiments. PLoS one 15(4): e0231137. https://doi.org/10.1371/journal.pone.0231137
27. Brenner E, Tiwari GR, Liu Y, Brock A, Mayfield RD. (2020) Single cell transcriptome profiling of the human alcohol-dependent brain. Human Mol Genet. ddaa038
26. Lele TP, Brock A, Peyton S. (2019) Emerging Concepts and Tools in Cell Mechanomemory. Ann Biomed Eng. https://doi.org/10.1007/s10439-019-02412-z
25. Johnson K, Howard G, Mo W, Strasser M, Lima, E ABF, Huang S, Brock A. (2019) Cancer cell population growth kinetics at low densities deviate from exponential growth model and suggest an Allee effect. PLoS Biol. 17(8): e3000399. 10.1371/journal.pbio.3000399.
24. Ma KY, Schonnesen AA, Brock A, Van Den Berg C, Eckhardt SG, Liu Z, Jiang N. (2019) Single-cell RNA sequencing of lung adenocarcinoma reveals heterogeneity of immune response–related genes. JCI Insight. 4(4):e 21387 (2019).
23. Jarrett AM, Lima E ABF, Hormuth DA, McKenna MT, Feng X, Ekrut DA, Resende AC, Brock A, Yankeelov TE. (2018) Mathematical models of tumor cell proliferation: A review of the literature. Expert Rev Anticancer Therapy 18 (12): 1271-1286. https://doi.org/10.1080/14737140.2018.1527689
22. Joyce MH, Lu C, James E, Hegab R, Allen S, Suggs L, Brock A. (2018) A Phenotypic Basis for Matrix Stiffness-Dependent Chemoresistance of Breast Cancer Cells to Doxorubicin. Front Oncol 8, 337, 10.3389/fonc.2018.00337
21. Al’Khafaji A, Deatherage D, Brock A. (2018) Control of Lineage-Specific Gene Expression by Functionalized gRNA Barcodes. ACS Synth Bio. 7 (10), 2468-2474. doi: 10.1021/acssynbio.8b00105
20. Howard GR, Johnson KE, Ayala AR, Yankeelov TE, Brock A. (2018) A multi-state model of chemoresistance to characterize phenotypic dynamics in breast cancer. Sci Rep 8 (1), 12058. https://doi.org/10.1038/s41598-018-30467-w
19. McKenna MT, Weis JA, Brock A, Quaranta V, Yankeelov TE. (2018) Precision Medicine with Imprecise Therapy: Computational Modeling for Chemotherapy in Breast Cancer. Translational Oncol 11 (3), 732-742. https://doi.org/10.1016/j.tranon.2018.03.009
38. Adler F, Anderson A.R.A., Bhushan A, Bogdan P, Bravo-Cordero JJ, Brock A, Chen Y, Cukierman E, DelGiorno KE, Denis GV, Ferrall-Fairbanks M, Gartner Z, Germain R, Gordon DM, Hunter G, Jolly MK, Karacosta LG, Mythreye K, Katira P, Kulkarni RP, Kutys ML, Lander AD, Laughney AM, Levine H, Lou E; Lowenstein PR, Masters KS, Pe'er D, Peyton SR, Platt MO, Purvis JE, Quon G, Richer JK, Riddle NC, Rodriguez A, Snyder JC, Szeto GL, Tomlin CJ, Yanai I, Zervantonakis I, Dueck HR. (2023) Modeling collective cell behavior in cancer: perspectives from an interdisciplinary conversation. Cell Systems. https://doi.org/10.1016/j.cels.2023.03.002
37. Howard G, Jost T, Yankeelov TE, Brock A. (2022) Quantification of long-term doxorubicin response dynamics in breast cancer cell lines to direct treatment schedules. PLOS Comp Biol 18 (3), e1009104. doi: https://doi.org/10.1101/2021.05.24.445407
36. Cotner M, Meng S, De Santiago C, Gardener A, Jost T, Brock A. (2022) Integration of quantitative methods and mathematical approaches for the modeling of cell proliferation dynamics. Amer. J. Cell Phys, 234(2), C247-262. doi.org/10.1152/ajpcell.00185.2022
35. Gardner A*, Morgan D*, Al’Khafaji A, Brock A. (2022) Functionalized Lineage Tracing for the Study and Manipulation of Heterogeneous Cell Populations. Biomedical Engineering Technologies. Methods in Molecular Biology, vol 2394. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-1811-0_8
34. Morgan D, Jost TA, DeSantiago C, Brock, A. (2021) Applications of high-resolution clone tracking technologies in cancer. Curr Opinion in Biomed Eng,100317.
33. Yang J, Virostko J, Hormuth DA, Liu J, Brock A, Kowalski J, Yankeelov TE (2021). An experimental-mathematical approach to predict tumor cell growth as a function of glucose availability in breast cancer cell lines. PloS one, 16 (7), e0240765.
32. Gutierrez C*, Al’Khafaji AM*, Brenner E*, Johnson KE, Gohil SH, Lin Z, Knisbacher BA, Durrett RE, Li S, Parvin S, Biran A, Zhang W, Rassenti L, Kipps TJ, Livak KJ, Neuberg D, Letai A, Getz G, Wu CJ, Brock A. (2021) Multifunctional barcoding with ClonMapper enables high-resolution study of clonal dynamics during tumor evolution and treatment. Nat Cancer, 2, 758–772. https://doi.org/10.1038/s43018-021-00222-8.
31. Liu J, Hormuth DA II, Davis T, Yang J, McKenna MT, Jarrett AM, Enderling H, Brock A, Yankeelov TE (2021) A time-resolved experimental–mathematical model for predicting the response of glioma cells to single-dose radiation therapy, Integrat Biol,13:7 167–183. https://doi.org/10.1093/intbio/zyab010
30. Kazerouni AS, Gadde M, Gardner A, Hormuth DA, Jarrett AM, Johnson KE, Lima EABF, Lorenzo G, Phillips C, Brock A, Yankeelov TE (2020). Integrating quantitative assays with biologically-based mathematical modeling for predictive oncology. iScience 101807. https://doi.org/10.1016/j.isci.2020.101807 3
29. Johnson K, Howard G, Morgan D, Brenner E, Gardner A, Durrett R, Mo W, Al’Khafaji A, Sontag E, Jarrett A, Yankeelov T, Brock A (2020) Integrating multimodal data sets into a mathematical framework to describe and predict therapeutic resistance in cancer. Phys Biol. 18 016001. https://doi.org/10.1088/1478-3975/abb09c.
28. Phillips CM, Lima EABF, Woodall RT, Brock A, Yankeelov TE (2020) A hybrid model of tumor growth and angiogenesis: In silico experiments. PLoS one 15(4): e0231137. https://doi.org/10.1371/journal.pone.0231137
27. Brenner E, Tiwari GR, Liu Y, Brock A, Mayfield RD. (2020) Single cell transcriptome profiling of the human alcohol-dependent brain. Human Mol Genet. ddaa038
26. Lele TP, Brock A, Peyton S. (2019) Emerging Concepts and Tools in Cell Mechanomemory. Ann Biomed Eng. https://doi.org/10.1007/s10439-019-02412-z
25. Johnson K, Howard G, Mo W, Strasser M, Lima, E ABF, Huang S, Brock A. (2019) Cancer cell population growth kinetics at low densities deviate from exponential growth model and suggest an Allee effect. PLoS Biol. 17(8): e3000399. 10.1371/journal.pbio.3000399.
24. Ma KY, Schonnesen AA, Brock A, Van Den Berg C, Eckhardt SG, Liu Z, Jiang N. (2019) Single-cell RNA sequencing of lung adenocarcinoma reveals heterogeneity of immune response–related genes. JCI Insight. 4(4):e 21387 (2019).
23. Jarrett AM, Lima E ABF, Hormuth DA, McKenna MT, Feng X, Ekrut DA, Resende AC, Brock A, Yankeelov TE. (2018) Mathematical models of tumor cell proliferation: A review of the literature. Expert Rev Anticancer Therapy 18 (12): 1271-1286. https://doi.org/10.1080/14737140.2018.1527689
22. Joyce MH, Lu C, James E, Hegab R, Allen S, Suggs L, Brock A. (2018) A Phenotypic Basis for Matrix Stiffness-Dependent Chemoresistance of Breast Cancer Cells to Doxorubicin. Front Oncol 8, 337, 10.3389/fonc.2018.00337
21. Al’Khafaji A, Deatherage D, Brock A. (2018) Control of Lineage-Specific Gene Expression by Functionalized gRNA Barcodes. ACS Synth Bio. 7 (10), 2468-2474. doi: 10.1021/acssynbio.8b00105
20. Howard GR, Johnson KE, Ayala AR, Yankeelov TE, Brock A. (2018) A multi-state model of chemoresistance to characterize phenotypic dynamics in breast cancer. Sci Rep 8 (1), 12058. https://doi.org/10.1038/s41598-018-30467-w
19. McKenna MT, Weis JA, Brock A, Quaranta V, Yankeelov TE. (2018) Precision Medicine with Imprecise Therapy: Computational Modeling for Chemotherapy in Breast Cancer. Translational Oncol 11 (3), 732-742. https://doi.org/10.1016/j.tranon.2018.03.009
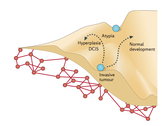
18. Brock A and Huang, S. (2017) Precision Oncology: Between Vaguely Right and Precisely Wrong. Cancer Research 77 (23), 6473-6479
17. Joyce MH, Allen S, Suggs L, Brock A. (2017) Novel Nanomaterials Enable Biomimetic Models of the Tumor Microenvironment. J. Nanotechnol. Article ID 5204163, https://doi.org/10.1155/2017/5204163.
16. Clark A, Bird N, Brock A. (2017) Intraductal Delivery to Rabbit Mammary Gland. J Vis Exp. 2017, 121:e 55209.
15. Brock A, Krause S, Ingber DE. (2015) Control of cancer formation by intrinsic genetic noise and microenviromental cues. Nat Rev Cancer, doi:10.1038/nrc3959.
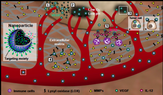
14. Kanapathipillai M, Brock A, Ingber D. Nanoparticle targeting of anti-cancer drugs that alter intracellular signaling or influence the tumor microenvironment. Adv Drug Delivery Rev 2014, 79, 107-118.
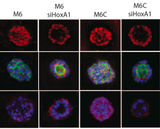
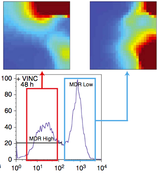
- 12. Pisco A*, Brock A*, Zhou J, Moor A, Mojtahedi M, Jackson D, Huang S. Non-Darwinian dynamics of therapy-induced cancer drug resistance (* equal contribution) Nat Comm 10.1038/ncomms3467, 2013) *= equal contribution
11. Krause S, Brock A, Ingber DE. Intraductal injection for localized delivery to the mouse mammary epithelium. J. Vis. Exp. e50692, doi:10.3791/50692, 2013.
10. Brock A, Goh HT, Yang B, Yu L, Li H, Loh YH. Cellular reprogramming: A new technology frontier in pharmaceutical research. Pharm Res. 29:35-52, 2011.
9. Brock A, Chang, H., Huang, S. Non-genetic heterogeneity – a mutation-independent driving force for somatic evolution of tumours. Nat Rev Genet, 10:336-42, 2009.
8. Brock AL, Ingber DE. Control of the direction of lamellipodia extension through changes in the balance between Rac and Rho activities. Mol Cell Biomech. 2(3):135-43, 2006.
7. Joy MP, Brock A, Ingber, DE, Huang S. High betweenness proteins in the yeast protein interaction network. J Biomed Biotechnol. 2:96-103, 2005.
6. Brock A, Huang, S., Ingber, DE. Identification of a distinct class of cytoskeleton-associated mRNAs using microarray technology. BMC Cell Biology. 4:6, 2003.
5. Brock A, Chang E, Ho CC, LeDuc, P, Jiang X, Whitesides GM, Ingber DE. Geometric determinants of directional cell motility revealed using microcontact printing. Langmuir. 19 (5); 1611-1617, 2003.
4. Parker KK, Brock AL, Brangwynne C, Mannix, RJ, Wang N, Ostuni E, Geisse, NA, Adams JC, Whitesides GM, Ingber DE. Directional control of lamellipodia extension by constraining cell shape and orienting cell tractional forces. FASEB. 16:1195-1204, 2002.
3. Hendricks CA, Razlog M, Matsuguchi T, Goyal A, Brock AL, Engelward BP. The S. cerevisiae Mag1 3-methyladenine DNA glycosylase modulates susceptibility to homologous recombination. DNA Repair. 1:645-659, 2002.
2. Bennett PV, Hada M, Hidema J, Lepre A, Pope LC, Quaite FE, Sullivan JH, Takayanagi S, Sutherland JC, Sutherland BM. (2001) Isolation of high molecular length DNA: alfalfa, pea, rice, sorghum, soybean and spinach. Crop Science 41, 167-172. doi:10.2135/cropsci2001.411167x
2. Bennett PV, Hada M, Hidema J, Lepre A, Pope LC, Quaite FE, Sullivan JH, Takayanagi S, Sutherland JC, Sutherland BM. (2001) Isolation of high molecular length DNA: alfalfa, pea, rice, sorghum, soybean and spinach. Crop Science 41, 167-172. doi:10.2135/cropsci2001.411167x
1. Lepre A, Sutherland JC, Trunk JG, Sutherland BM. A robust, inexpensive filter for blocking UVC radiation in broad-spectrum ‘UVB’ lamps. J Photochem Photobiol B. 43(1):34-40, 1998.
