Brock Lab
Our lab investigates the role of heterogeneity in cell state transitions, cancer progression, and therapeutic responses.
|
Our Focus
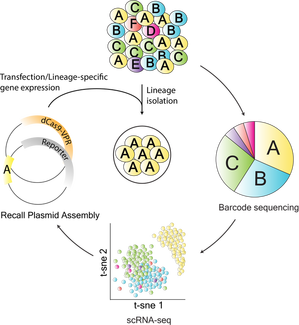
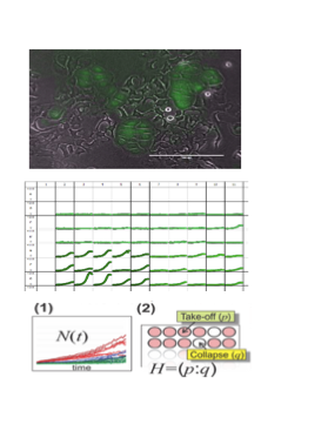
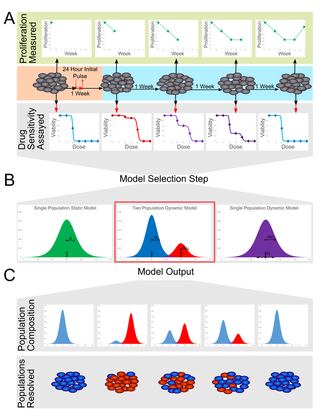
Drug resistance of tumor cells is the cause of treatment failure for most human cancers. Acquired resistance was observed in the very first clinical administration of a chemotherapeutic agent in 1942. Since that time, vast improvements in chemotherapy- including novel drugs, combination therapies, reduced toxicity, and highly targeted treatments- have extended the lives of many cancer patients. Yet drug resistance remains a major scientific and clinical challenge in the treatment of human cancer. Tumor cell populations are characterized by enormous heterogeneity and plasticity. This variation contributes to disease progression and response to therapy, although the mechanisms are not yet well understood. To address these challenges, we are developing novel technologies and experimental systems rooted in the tools of systems and synthetic biology and bioengineering. |
Lab News
04/20/2023 Amy is a keynote speaker at the CPRIT Symposium on "Expanding Texas Leadership in Computational Oncology throughout the Cancer Continuum" today at Dell Medical School, UT Austin. 04/13/2023 Amy will be giving a talk and Carolina, Daylin, Didi, and Tyler will present posters at AACR 2023. Come visit with us! 02/2023 Congratulations to Didi on receiving the NOA for her F31 fellowship! 09/2023 The lab welcomes new graduate student Audrey Freischel! 08/2022 Congratulations to Sarah Meng, Michael Cotner and Kennedy Howland on passing their BME qualifying exams! |